Modeling Pathogenic Variants in the RNA Exosome
DOI: 10.14800/rd.1166
Abstract
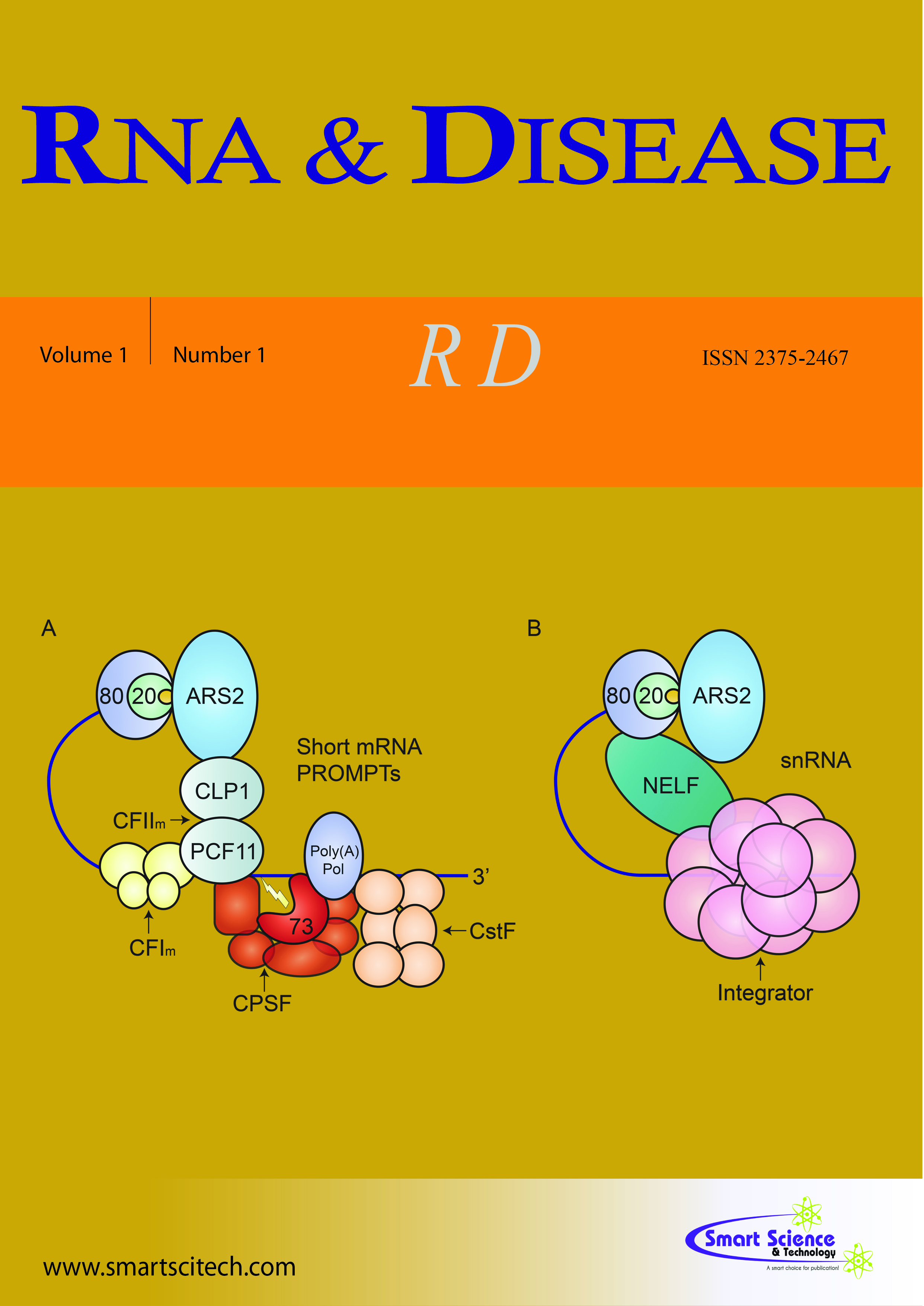
Exosomopathies are a collection of rare diseases caused by mutations in genes that encode structural subunits of the RNA exosome complex. The RNA exosome is critical for both processing and degrading many RNA targets. Mutations in individual RNA exosome subunit genes (termed EXOSC genes) are linked to a variety of distinct diseases. These exosomopathies do not arise from homozygous loss-of-function or large deletions in the EXOSC genes likely because some level of RNA exosome activity is essential for viability. Thus, all patients described so far have at least one allele with a missense mutation encoding an RNA exosome subunit with a single pathogenic amino acid change linked to disease. Understanding how these changes lead to the disparate clinical presentations that have been reported for this class of diseases necessitates investigation of how individual pathogenic missense variants alter RNA exosome function. Such studies will require access to patient samples, a challenge for these very rare diseases, coupled with modeling the patient variants. Here, we highlight five recent studies that model pathogenic variants in EXOSC3, EXOSC2, and EXOSC5.